Research
- English
- Türkçe
In eukaryotic cells, DNA usually wrapped around histone proteins. This structure is called a nucleosome. Nucleosomes are fundamental units of chromatin and it packages long DNA molecules into a more compact shape. During the cell cycle, chromatin undergoes various structural changes. Histone proteins can be modified with different post-translational modifications that alter chromatin structure. These modifications can affect gene expression. For example, some modifications loosening the structure which increases the accessibility of the chromatin for replication or translation. On the other hand, some other modifications decrease the accessibility which causes transcription repression and compaction. Even if all nucleated cells in an organism share the same DNA pattern which originates from totipotent zygote these modifications alter gene expression in different cells of the organism and this causes lineage differentiation and affects cell fate. During development, embryonic stem cells differentiate into three major lineages which are ectoderm, mesoderm, and endoderm.
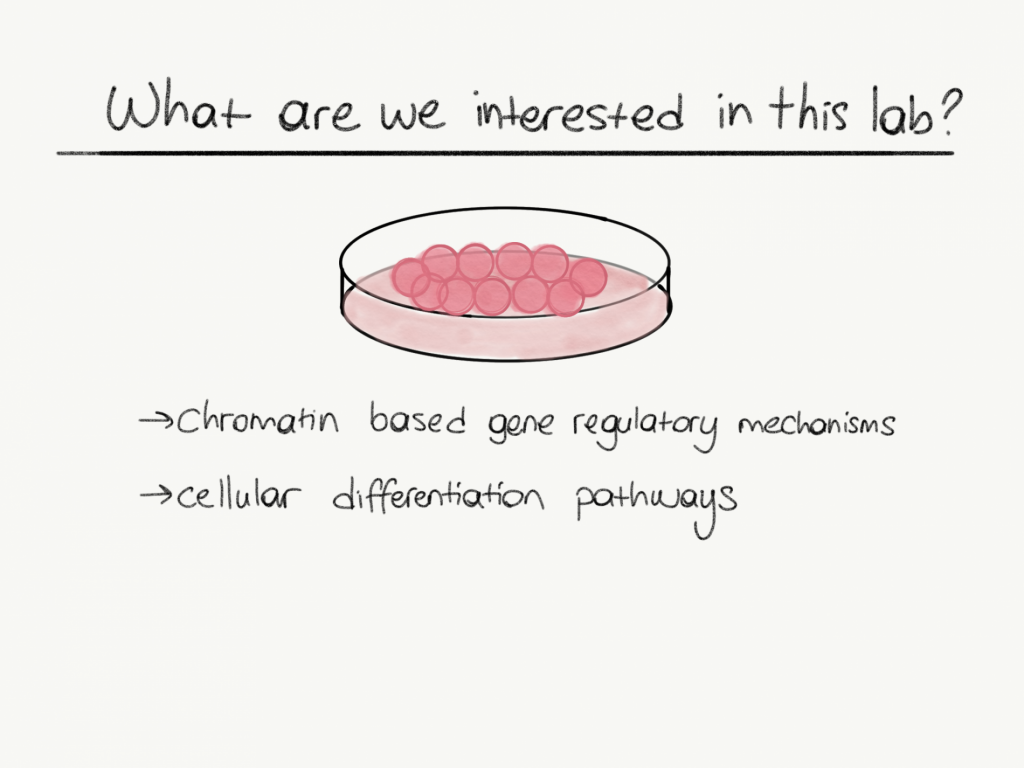
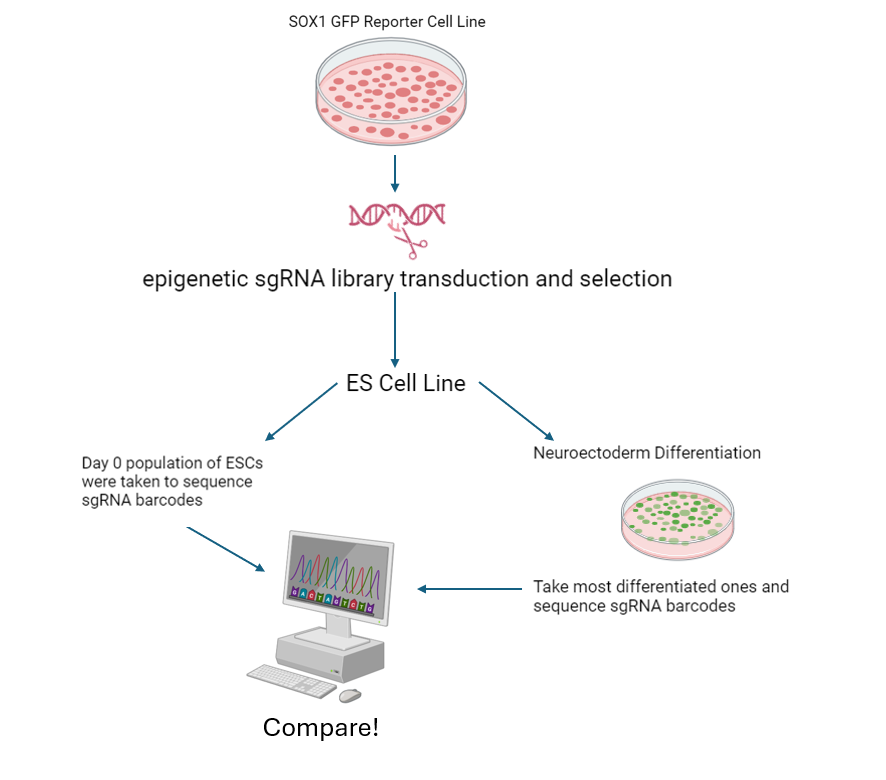
In our lab, we focused on epigenetic control of cell fate decision mechanisms using mouse embryonic stem cells as a model. Our main interest is epigenetic factors that affecting nucleosome remodeling, chromatin dynamics and their effect on cell fate decisions via gene expression regulation.
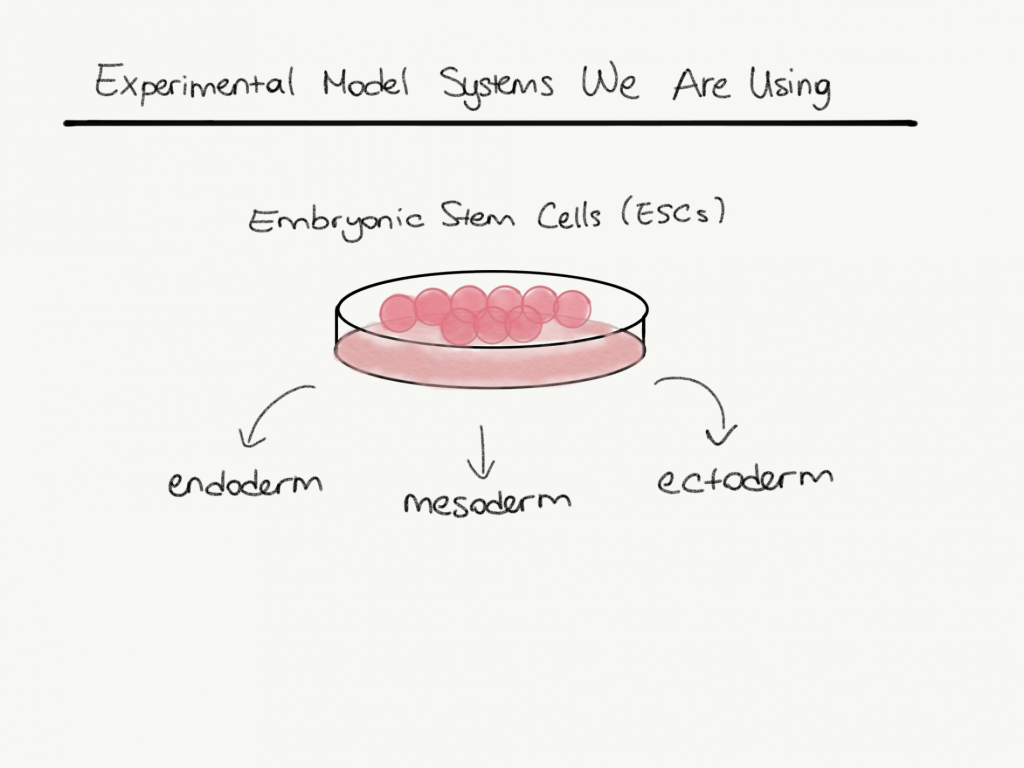
We use mouse embryonic stem cell cultures, reporter cell lines, genetic manipulation, non-coding RNAs, proteomics and transcriptomics analysis, and protein-protein interaction assays in our lab.
Currently, we are investigating several proteins and protein complexes’ interactions with each other and with chromatin to understand their effects on different pathways that affecting pluripotency, self-renewal, and cell fate decision.